DC CRISPR Initiative at HacDC: Difference between revisions
From HacDC Wiki
(Created page with "==Project Description== CRISPR-Cas9 is a groundbreaking new gene editing technique in molecular biology. In brief, ten years ago it was found that bacteria possess a self-def...") |
(→Sample Inventory: asdf) |
||
| (29 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
==Project Description== | ==Project Description== | ||
CRISPR-Cas9 is a groundbreaking new gene editing technique in | * Organizers: Enrique C., Nancy C. Wolfson, Bobby B. | ||
* Contact: [email protected] | |||
CRISPR-Cas9 is a groundbreaking new (2012 vintage) gene editing technique. While gene editing is not a new concept, previous methods were far more expensive, slow and restricted in capabilities than CRISPR. Further, whereas previous methods only successfully edited a few percent of the exposed cells, CRISPR's efficiency approaches 100%. This is particularly important for gene editing in living multi-cellular organisms. The new technique is dramatically accelerating the pace of genetic engineering since its invention in 2012. | |||
CRISPR is an acronym that describes a genetic curiosity observed several decades ago: Clusters of Regularly Interspaced Short Palindromic Repeats. A few years go it was recognized that these odd DNA sequences in bacteria are deactivated virus DNA and make up a kind of Virus Definition Database. Finally, it was discovered that a protein exists which goes around scanning bacteria DNA searching for matches to virus DNA sequences and, when a match is found, cuts the gene in the bacteria DNA very precisely and efficiently. This protein was termed Cas9. In effect, bacteria and other organisms already have an excellent built-in gene-editing mechanism, and scientists have since learned to hijack that mechanism using engineered RNA sequences that direct the Cas9 protein to cut DNA in any desired location in the genome. | |||
The Cas9 mechanism only cuts the DNA in one location, leaving DNA repair mechanisms to fix the double-strand cut. Repair mechanisms for such a serious double-strand break are so imperfect as to incapacitate the gene with errors most of the time. Thus the Cas9 protein deactivates genes rather than removing them. However, by programming two custom RNA target strands, two Cas9 proteins can be used in tandem to excise part of a genome altogether. To add a new gene, there must be enough of that gene floating around during the CRISPR process that it becomes incorporated into the genome via the repair mechanisms. | |||
These are the best videos we've found so far: | |||
Genome Editing with CRISPR-Cas9 by McGovern Institute for Brain Research | |||
https://www.youtube.com/watch?v=2pp17E4E-O8 | |||
What is CRISPR? by Bozeman Science | |||
https://www.youtube.com/watch?v=MnYppmstxIs | |||
Read more here: | |||
https://en.wikipedia.org/wiki/CRISPR | |||
Read more here: | Read more here: | ||
https://en.wikipedia.org/wiki/CRISPR | https://en.wikipedia.org/wiki/CRISPR | ||
In the ODIN kit experiment, bacteria (E. coli HME63 strain) are modified to add resistance to the antibiotic streptomycin. The kit provides the vulnerable bacteria, the resistance gene, and growth media with and without antibiotic. The original unmodified bacteria can only grow on the plain agar media whereas bacteria with a successfully edited genome will also grow on the streptomycin-laced agar. | In the ODIN kit experiment, bacteria (E. coli HME63 strain) are modified to add resistance to the antibiotic streptomycin. The kit provides the vulnerable bacteria, the resistance gene, and growth media with and without antibiotic. The original unmodified bacteria can only grow on the plain agar media whereas bacteria with a successfully edited genome will also grow on the streptomycin-laced agar. This is similar to Wenyan Jiang, David Bikard, David Cox, Feng Zhang and Luciano Marraffini, RNA-guided editing of bacterial genomes using CRISPR-Cas systems, Nature Biotechnology 31(3), pp.233 (2013). http://zlab.mit.edu/assets/reprints/Jiang_W_Nat_Biotechnol_2013.pdf | ||
==Financial Support / Sponsors== | |||
HacDC - 8/2016 - purchased Bacterial DIY CRISPR kit. | |||
Enrique C - 11/2016- purchased Bacterial DIY CRISPR Kit. | |||
Nancy W - purchased Bacterial DIY CRISPR kit. | |||
Enrique C - 5/2017 - purchased Bacterial DIY CRISPR Kit Refill. | |||
Nancy W - 2017 - purchased Bacterial DIY CRISPR kit refill. | |||
Nancy W - 2017 - purchased temperature-controlled water bath. | |||
Nancy W - 2017 - loaned 1600X Optical Microscope with USB camera. | |||
Enrique C - 2017 - purchased supplies and gram staining kit. | |||
The ODIN - 01/2018- provided free sample of next-gen CRISPR kit. | |||
==Activities and Goals== | ==Activities and Goals== | ||
| Line 14: | Line 42: | ||
Enrique C. - Project Manager and Point of Contact | Enrique C. - Project Manager and Point of Contact | ||
Nancy W. - Project Development Lead | Nancy W. - Project Development Lead | ||
Bobby B. - Molecular Biology Advisor | |||
Sophia M. - Scientific Advisor | |||
==Worklog== | ==Worklog== | ||
| Line 28: | Line 57: | ||
August 10, 2016 | August 10, 2016 | ||
Ken, Bobby, Nancy, and Enrique. We streaked some of the original E. coli HME63 bacteria onto two plain agar plates. Plate 1 was left out tonight (the bacteria need to grow). Plate 2 was immediately refrigerated and will be taken out to grow just before the actual experiment. | Ken, Bobby, Nancy, and Enrique. We streaked some of the original E. coli HME63 bacteria onto two plain agar plates. Plate 1 was left out tonight (the bacteria need to grow). Plate 2 was immediately refrigerated and will be taken out to grow just before the actual experiment. | ||
August, 2016 | |||
Nancy, Ken and Enrique. We performed the CRISPR experiment, but realized we don't have a constant-temperature water bath. We used an IR thermometer and the microwave to prepare and maintain a water bath of approximately 42C. The incubation period post-CRISPR is also quite long, up to 4 hours at room temperature, which makes this experiment problematic as an after-work evening activity. It'd work better on a weekend day or holiday since we have day jobs. | |||
September, 2016 | |||
Nancy, Enrique. The first CRISPR-modified bacteria on the Strep-Kan plate was left at room temperature over the weekend (48 hours). However, no bacteria colonies could be easily seen in the plate. The plate was left at room temperature several more days with no change. It looks like our first attempt did not wildly succeed. | |||
September, 2016 | |||
Enrique. I prepared a new set of Agar (3) and Strep-Kan Agar (3) plates for troubleshooting experiments and a second try at CRISPR. We should test the full CRISPR protocol again, both on Agar and Strep-Kan plates, but also test the survival of bacteria at several other steps: Transformation Mix only and Transformation Mix + tracrRNA + crRNA. | |||
September, 2016 | |||
Enrique and Nancy. We developed a troubleshooting protocol under the suspicion that maybe none of the bacteria are surviving (we did not make a plain Agar control plate last time). We also tested the 'sterile' innoculation loops vs. a bag of zip-ties we found laying around. Nancy brought an alcohol thermometer that's probably more accurate in water than the IR thermometer. The resulting plates were incubated for 24-48 hours. It was difficult to re-suspend the bacteria in solution after harvesting from the first plate. A vortex generator (basically a strong vibrator) would be helpful. | |||
September, 2016 | |||
Enrique and Nancy. Hm... there are some specs on the CRISPR plate this time, just 2-3 colonies though. All the other (Agar) plates had tons of bacteria, so clearly the low efficiency is at the CRISPR step with the Template DNA. Also we brought in more Bleach for disposal of old samples, and bleach wipes. | |||
September, 2016 | |||
Enrique. I located a much better (real) biological microscope with up to 1000x magnification in the basement. Man our inventory sucks; I had no idea we had this thing. Images are much better, although I'm not certain we're looking at individual bacteria. I also purchased a hotplate so we can actually control temperature baths in the future. | |||
January, 2017 | |||
See notebook. | |||
February, 2017. | |||
The third trial of the CRISPR process gave us a positive control sample, which makes the result inconclusive. While the Agar media grew more, both the CRISPR and original LD 21 bacteria grew somewhat on the Strep-Kan media. We perfomed a second control experiment, plating 100uL of the LD21 bacteria and the Transformation-Mix HME 63 strain with 400uL of DI water each, onto Strep-Kan media. | |||
June, 2017 | |||
Purchased Bacterial CRISPR refill kit using HacDC Project CRISPR funds. | |||
July 2017 - Bobby, Nancy, Enrique, Sophia and Richard. | |||
Met to plan workshop. Bobby gave us his Introduction to Molecular Biology talk - it was about 90 minutes but could probably be shortened to 60 if needed. Prepared fresh Agar and Strep-Kan plates. Sophia showed us a new (correct) streaking technique. We plated some bacteria but they overgrew (storage place on top of the fridge is too warm in the summer), making it impossible to isolated a colony. Sophia took a second batch home and it was better with some isolated colonies. | |||
August 23, 2017 - Sophia, Nancy and Enrique. | |||
Trial 4 of the full CRISPR process. Enrique took the resulting plates home to monitor growth at room temperature. No growth observed on the CRISPR sample. Control sample grew quickly. | |||
December 11, 2017 - Nancy and Enrique. | |||
Contacted kit supplier to obtain fresh ingredients. | |||
January 09, 2017 - Enrique. | |||
Prepared fresh Agar and Strep-Kan Agar plates. Checked for contamination growth 48 hours (none). | |||
January 12, 2017 - Nancy, Enrique and guests Tobi and Tom. | |||
Trial 4.5 of the CRISPR process. Used bacteria straight from the supply rather than a colony (not available). The result after 48 hours was much growth on the Agar plate and zero growth on the Strep-Kan plate. (no modification) | |||
== Sample Inventory == | |||
'''CRISPR Trial 8/24/2017 | |||
''' | |||
LB-Agar Plate at 1 and 20 Hours: | |||
[[File:IMG_20170825_001628s.jpg]] [[File:IMG_20170825_190805s.jpg]] | |||
StrepKan Plate at 1, 20, 28 and 57 Hours: | |||
[[File:IMG_20170825_001648s.jpg]] [[File:IMG_20170825_190912s.jpg]] [[File:IMG_20170826_032748s.jpg]] [[File:IMG_20170827_093620s.jpg]] | |||
'''CRISPR Trial 1/12/2018 | |||
'''Clean Agar plate after 48 hours incubation. No contamination growth observed. | |||
[[Media:CRISPR-180112-Agar_blank_48h.jpeg]] | |||
Clean Strep-Kan plate after 48 hours incubation. No contamination growth observed. | |||
[[Media:CRISPR-180112-StrepKan_blank_48h.jpeg]] | |||
Agar plate with the post-protocol bacteria incubated 48 hours (control sample): much growth (overgrown). | |||
[[Media:CRISPR-180112-Agar_Bact_48h.jpeg]] | |||
Strep-Kan plate with the post-protocol bacteria incubation 48 hours (CRISPR result): no growth. | |||
[[Media:CRISPR-180112-StrepKan_CrisprBact_48h.jpeg]] | |||
== News References == | |||
* Video: bacteria evolving antibiotic resistance in 11 days. https://www.youtube.com/watch?v=yybsSqcB7mE | |||
* Video: CRISPR Patent Controversy: https://www.youtube.com/watch?v=IboHEQumDGc | |||
Latest revision as of 02:08, 27 January 2018
Project Description
- Organizers: Enrique C., Nancy C. Wolfson, Bobby B.
- Contact: [email protected]
CRISPR-Cas9 is a groundbreaking new (2012 vintage) gene editing technique. While gene editing is not a new concept, previous methods were far more expensive, slow and restricted in capabilities than CRISPR. Further, whereas previous methods only successfully edited a few percent of the exposed cells, CRISPR's efficiency approaches 100%. This is particularly important for gene editing in living multi-cellular organisms. The new technique is dramatically accelerating the pace of genetic engineering since its invention in 2012.
CRISPR is an acronym that describes a genetic curiosity observed several decades ago: Clusters of Regularly Interspaced Short Palindromic Repeats. A few years go it was recognized that these odd DNA sequences in bacteria are deactivated virus DNA and make up a kind of Virus Definition Database. Finally, it was discovered that a protein exists which goes around scanning bacteria DNA searching for matches to virus DNA sequences and, when a match is found, cuts the gene in the bacteria DNA very precisely and efficiently. This protein was termed Cas9. In effect, bacteria and other organisms already have an excellent built-in gene-editing mechanism, and scientists have since learned to hijack that mechanism using engineered RNA sequences that direct the Cas9 protein to cut DNA in any desired location in the genome.
The Cas9 mechanism only cuts the DNA in one location, leaving DNA repair mechanisms to fix the double-strand cut. Repair mechanisms for such a serious double-strand break are so imperfect as to incapacitate the gene with errors most of the time. Thus the Cas9 protein deactivates genes rather than removing them. However, by programming two custom RNA target strands, two Cas9 proteins can be used in tandem to excise part of a genome altogether. To add a new gene, there must be enough of that gene floating around during the CRISPR process that it becomes incorporated into the genome via the repair mechanisms.
These are the best videos we've found so far:
Genome Editing with CRISPR-Cas9 by McGovern Institute for Brain Research
https://www.youtube.com/watch?v=2pp17E4E-O8
What is CRISPR? by Bozeman Science https://www.youtube.com/watch?v=MnYppmstxIs
Read more here: https://en.wikipedia.org/wiki/CRISPR
Read more here: https://en.wikipedia.org/wiki/CRISPR
In the ODIN kit experiment, bacteria (E. coli HME63 strain) are modified to add resistance to the antibiotic streptomycin. The kit provides the vulnerable bacteria, the resistance gene, and growth media with and without antibiotic. The original unmodified bacteria can only grow on the plain agar media whereas bacteria with a successfully edited genome will also grow on the streptomycin-laced agar. This is similar to Wenyan Jiang, David Bikard, David Cox, Feng Zhang and Luciano Marraffini, RNA-guided editing of bacterial genomes using CRISPR-Cas systems, Nature Biotechnology 31(3), pp.233 (2013). http://zlab.mit.edu/assets/reprints/Jiang_W_Nat_Biotechnol_2013.pdf
Financial Support / Sponsors
HacDC - 8/2016 - purchased Bacterial DIY CRISPR kit. Enrique C - 11/2016- purchased Bacterial DIY CRISPR Kit. Nancy W - purchased Bacterial DIY CRISPR kit. Enrique C - 5/2017 - purchased Bacterial DIY CRISPR Kit Refill. Nancy W - 2017 - purchased Bacterial DIY CRISPR kit refill. Nancy W - 2017 - purchased temperature-controlled water bath. Nancy W - 2017 - loaned 1600X Optical Microscope with USB camera. Enrique C - 2017 - purchased supplies and gram staining kit. The ODIN - 01/2018- provided free sample of next-gen CRISPR kit.
Activities and Goals
DC CRISPR Initiative is our effort to learn about, perform, and teach CRISPR genetic editing at HacDC. To begin the project, we’ve ordered a Do-It-Yourself CRISPR Kit, which includes (supposedly) all the tools and ingredients needed to perform a CRISPR procedure a few times. We’ll hold a few events at HacDC to go through the procedure and document our experience. Eventually we’ll create a guide that older high school kids can follow. This project also explores interest in molecular biology and genetics at HacDC. We're just starting! Keep an eye out for CRISPR events in our MeetUp page, on the mailing list, and our Blabber discussion forum.
Project Team Members
Enrique C. - Project Manager and Point of Contact Nancy W. - Project Development Lead Bobby B. - Molecular Biology Advisor Sophia M. - Scientific Advisor
Worklog
July 30, 2016 We received the CRISPR kit purchased with Project EXPANSION funds (thanks!).
August 2, 2016 Nancy and Enrique inventoried the ODIN kit and designated the small classroom fridge as the "NO FOOD" Project CRISPR fridge.
August 5, 2016 Nancy and Enrique prepared four Petri dishes (two plain agar, two streptomycin-agar). The agar and antibiotic(streptomycin)-laced agar are gel-like substances similar to gelatin. They come as powders which must be mixed with water and heated to dissolve. The recipe is proportioned for seven Petri dishes but we scaled down to one of each, scaling the agar powders and water by one-seventh. Even so we were able to coat two dishes with each growth medium. We didn't have distilled or deionized water and used bottled purified drinking water in a pinch. The mixture (agar gel only, no bacteria!) was heated in the microwave 7 seconds at a time. It took 4-5 cycles until the powders were fully dissolved and the liquid transparent, then another 5 minutes until they were cool enough to handle and pour into the plastic Petri dishes. The dishes cooled at room temperature for an hour to remove some condensation (the covered hot liquid creates condensation on the lid), then placed in the fridge. Two are agar (no antibiotic) and two are streptomycin/Kan agar (antibiotic laced).
August 10, 2016 Ken, Bobby, Nancy, and Enrique. We streaked some of the original E. coli HME63 bacteria onto two plain agar plates. Plate 1 was left out tonight (the bacteria need to grow). Plate 2 was immediately refrigerated and will be taken out to grow just before the actual experiment.
August, 2016 Nancy, Ken and Enrique. We performed the CRISPR experiment, but realized we don't have a constant-temperature water bath. We used an IR thermometer and the microwave to prepare and maintain a water bath of approximately 42C. The incubation period post-CRISPR is also quite long, up to 4 hours at room temperature, which makes this experiment problematic as an after-work evening activity. It'd work better on a weekend day or holiday since we have day jobs.
September, 2016 Nancy, Enrique. The first CRISPR-modified bacteria on the Strep-Kan plate was left at room temperature over the weekend (48 hours). However, no bacteria colonies could be easily seen in the plate. The plate was left at room temperature several more days with no change. It looks like our first attempt did not wildly succeed.
September, 2016 Enrique. I prepared a new set of Agar (3) and Strep-Kan Agar (3) plates for troubleshooting experiments and a second try at CRISPR. We should test the full CRISPR protocol again, both on Agar and Strep-Kan plates, but also test the survival of bacteria at several other steps: Transformation Mix only and Transformation Mix + tracrRNA + crRNA.
September, 2016 Enrique and Nancy. We developed a troubleshooting protocol under the suspicion that maybe none of the bacteria are surviving (we did not make a plain Agar control plate last time). We also tested the 'sterile' innoculation loops vs. a bag of zip-ties we found laying around. Nancy brought an alcohol thermometer that's probably more accurate in water than the IR thermometer. The resulting plates were incubated for 24-48 hours. It was difficult to re-suspend the bacteria in solution after harvesting from the first plate. A vortex generator (basically a strong vibrator) would be helpful.
September, 2016 Enrique and Nancy. Hm... there are some specs on the CRISPR plate this time, just 2-3 colonies though. All the other (Agar) plates had tons of bacteria, so clearly the low efficiency is at the CRISPR step with the Template DNA. Also we brought in more Bleach for disposal of old samples, and bleach wipes.
September, 2016 Enrique. I located a much better (real) biological microscope with up to 1000x magnification in the basement. Man our inventory sucks; I had no idea we had this thing. Images are much better, although I'm not certain we're looking at individual bacteria. I also purchased a hotplate so we can actually control temperature baths in the future.
January, 2017
See notebook.
February, 2017. The third trial of the CRISPR process gave us a positive control sample, which makes the result inconclusive. While the Agar media grew more, both the CRISPR and original LD 21 bacteria grew somewhat on the Strep-Kan media. We perfomed a second control experiment, plating 100uL of the LD21 bacteria and the Transformation-Mix HME 63 strain with 400uL of DI water each, onto Strep-Kan media.
June, 2017 Purchased Bacterial CRISPR refill kit using HacDC Project CRISPR funds.
July 2017 - Bobby, Nancy, Enrique, Sophia and Richard. Met to plan workshop. Bobby gave us his Introduction to Molecular Biology talk - it was about 90 minutes but could probably be shortened to 60 if needed. Prepared fresh Agar and Strep-Kan plates. Sophia showed us a new (correct) streaking technique. We plated some bacteria but they overgrew (storage place on top of the fridge is too warm in the summer), making it impossible to isolated a colony. Sophia took a second batch home and it was better with some isolated colonies.
August 23, 2017 - Sophia, Nancy and Enrique. Trial 4 of the full CRISPR process. Enrique took the resulting plates home to monitor growth at room temperature. No growth observed on the CRISPR sample. Control sample grew quickly.
December 11, 2017 - Nancy and Enrique. Contacted kit supplier to obtain fresh ingredients.
January 09, 2017 - Enrique. Prepared fresh Agar and Strep-Kan Agar plates. Checked for contamination growth 48 hours (none).
January 12, 2017 - Nancy, Enrique and guests Tobi and Tom. Trial 4.5 of the CRISPR process. Used bacteria straight from the supply rather than a colony (not available). The result after 48 hours was much growth on the Agar plate and zero growth on the Strep-Kan plate. (no modification)
Sample Inventory
CRISPR Trial 8/24/2017
LB-Agar Plate at 1 and 20 Hours:
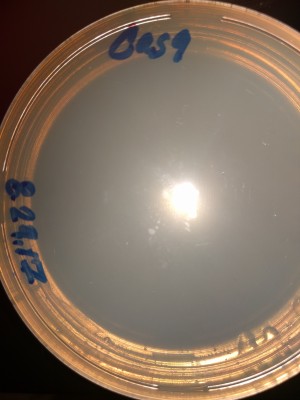
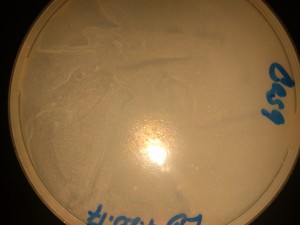
StrepKan Plate at 1, 20, 28 and 57 Hours:
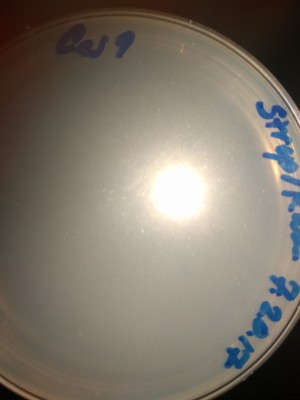

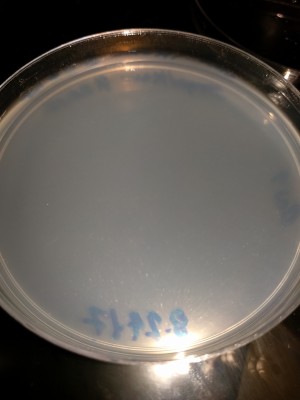
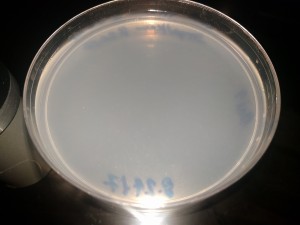
CRISPR Trial 1/12/2018 Clean Agar plate after 48 hours incubation. No contamination growth observed. Media:CRISPR-180112-Agar_blank_48h.jpeg Clean Strep-Kan plate after 48 hours incubation. No contamination growth observed. Media:CRISPR-180112-StrepKan_blank_48h.jpeg Agar plate with the post-protocol bacteria incubated 48 hours (control sample): much growth (overgrown). Media:CRISPR-180112-Agar_Bact_48h.jpeg Strep-Kan plate with the post-protocol bacteria incubation 48 hours (CRISPR result): no growth. Media:CRISPR-180112-StrepKan_CrisprBact_48h.jpeg
News References
- Video: bacteria evolving antibiotic resistance in 11 days. https://www.youtube.com/watch?v=yybsSqcB7mE
- Video: CRISPR Patent Controversy: https://www.youtube.com/watch?v=IboHEQumDGc
